Diagnostic of assessments in MSE: did Assess models converge during MSE?
Source:R/diagnostic.R
diagnostic.RdDiagnostic check for convergence of Assess models during closed-loop simulation. Use when the MP was
created with make_MP with argument diagnostic = "min" or "full".
This function summarizes and plots the diagnostic information.
Arguments
- MSE
An object of class MSE created by
MSEtool::runMSE().- MP
Optional, a character vector of MPs that use assessment models.
- gradient_threshold
The maximum magnitude (absolute value) desired for the gradient of the likelihood.
- figure
Logical, whether a figure will be drawn.
- ...
Arguments to pass to
diagnostic.
Value
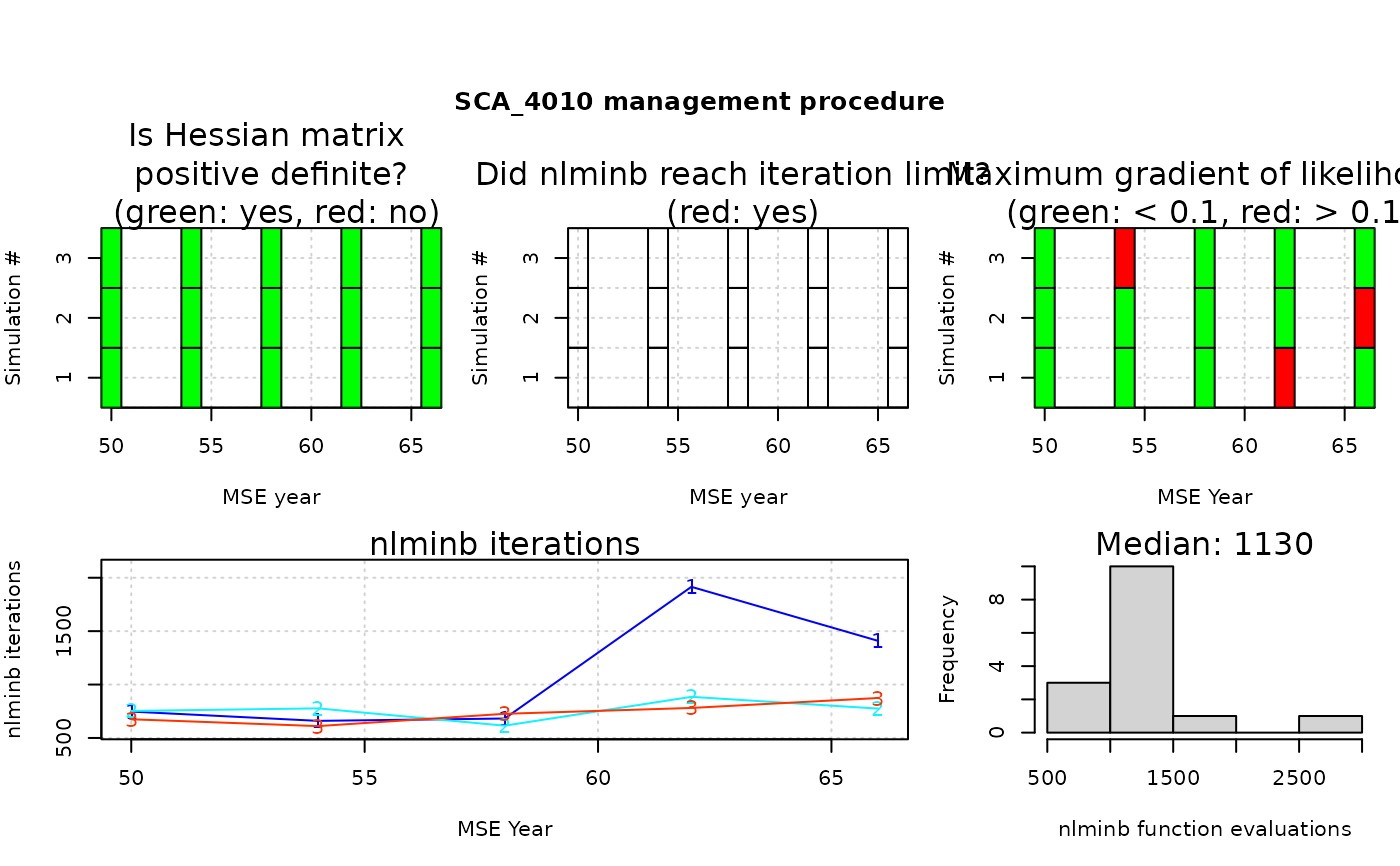
A matrix with diagnostic performance of assessment models in the MSE. If figure = TRUE,
a set of figures: traffic light (red/green) plots indicating whether the model converged (defined if a positive-definite
Hessian matrix was obtained), the optimizer reached pre-specified iteration limits (as passed to stats::nlminb()),
and the maximum gradient of the likelihood in each assessment run. Also includes the number of optimization iterations
function evaluations reported by stats::nlminb() for each application of the assessment model.
Examples
# \donttest{
OM <- MSEtool::testOM; OM@proyears <- 20
myMSE <- runMSE(OM, MPs = "SCA_4010")
#> → Loading operating model
#> → Calculating MSY reference points for each year
#> → Optimizing for user-specified depletion in last historical year
#> → Calculating historical stock and fishing dynamics
#> → Calculating per-recruit reference points
#> → Calculating B-low reference points
#> → Calculating reference yield - best fixed F strategy
#> → Simulating observed data
#> ✔ Running forward projections
#>
#> → 1/1 Running MSE for: "SCA_4010"
#>
|=== | 5 % ~21s
|====== | 11% ~10s
|======== | 16% ~06s
|=========== | 21% ~05s
|============== | 26% ~07s
|================ | 32% ~05s
|=================== | 37% ~04s
|====================== | 42% ~04s
|======================== | 47% ~05s
|=========================== | 53% ~04s
|============================= | 58% ~03s
|================================ | 63% ~02s
|=================================== | 68% ~03s
|===================================== | 74% ~02s
|======================================== | 79% ~02s
|=========================================== | 84% ~01s
|============================================= | 89% ~01s
|================================================ | 95% ~00s
|==================================================| 100% elapsed=08s
#>
diagnostic(myMSE)
#> ✔ Creating plots for MP:
#> SCA_4010
 #> SCA_4010
#> Percent positive-definite Hessian 100
#> Percent iteration limit reached 0
#> Percent max. gradient < 0.1 100
#> Median iterations 777
#> Median function evaluations 1130
# How to get all the reporting
library(dplyr)
#>
#> Attaching package: ‘dplyr’
#> The following objects are masked from ‘package:stats’:
#>
#> filter, lag
#> The following objects are masked from ‘package:base’:
#>
#> intersect, setdiff, setequal, union
conv_statistics <- lapply(1:myMSE@nMPs, function(m) {
lapply(1:myMSE@nsim, function(x) {
myMSE@PPD[[m]]@Misc[[x]]$diagnostic %>%
mutate(MP = myMSE@MPs[m], Simulation = x)
}) %>% bind_rows()
}) %>% bind_rows()
# }
#> SCA_4010
#> Percent positive-definite Hessian 100
#> Percent iteration limit reached 0
#> Percent max. gradient < 0.1 100
#> Median iterations 777
#> Median function evaluations 1130
# How to get all the reporting
library(dplyr)
#>
#> Attaching package: ‘dplyr’
#> The following objects are masked from ‘package:stats’:
#>
#> filter, lag
#> The following objects are masked from ‘package:base’:
#>
#> intersect, setdiff, setequal, union
conv_statistics <- lapply(1:myMSE@nMPs, function(m) {
lapply(1:myMSE@nsim, function(x) {
myMSE@PPD[[m]]@Misc[[x]]$diagnostic %>%
mutate(MP = myMSE@MPs[m], Simulation = x)
}) %>% bind_rows()
}) %>% bind_rows()
# }